Rapid screening for probiotic bacteria



In this series of articles, Will Chu, Science Editor for NutraIngredients, discusses some of the key issues and challenges facing the nutraceutical and food ingredient industry today.
As consumer demand for ‘healthy’ products rises, there is ever-increasing interest in finding novel probiotics. However, the right organism has to be identified from thousands of potential bacterial candidates. Robyn Eijlander, Molecular Microbiology expert from NIZO, explains how computational screening models can speed up the identification of potential probiotics, enabling food manufacturers to bring products to market more quickly and cost-effectively.
Robyn Eijlander: While there are many probiotics already on the market, food manufacturers might want to differentiate their product with a novel probiotic. Perhaps they need a probiotic that better fits with their desired processes or type of product: one that is easier to culture or faster to grow. Or they might want to find a benefit or application that isn’t yet on the market. Conversely, they might have a large bacterial culture collection for which they want to add commercial value.
RE: There are many possible approaches. You can test ‘good’ bacteria commonly found on (for instance) human skin or in the gut for probiotic traits. Or you can follow-up on the results of existing clinical studies that demonstrated a health benefit from a food or compound, to see if you can identify a bacterium that may be associated with that benefit. Traditional foods that are known within their communities to be ‘healthy’ can be a good source of new probiotics. One example is natto, traditional fermented Japanese soybeans: research discovered that the bacterial strain used in the fermentation, Bacillus subtills natto, produces vitamin K2.
You can also start with a known probiotic strain and try to find others in the same species that are also probiotics. For example, Lacticaseibacillus rhamnosus (previously Lactobacillus rhamnosus)GG (LGG) is a widely used probiotic, so you could compare other L. rhamnosus strains, or even related lactic acid bacteria (LAB) to see if any of them are also probiotic. Yet another approach is to start with computational genome screening to find bacterial functionalities that are known to exert a certain probiotic trait.
RE: Substantiating the health benefits of a potential probiotic using in vitro and in vivo studies is expensive and can take a long time, especially if you don’t know the potential molecular mechanisms behind the benefit. This is even more so when you have hundreds of potential candidate strains. You need to reduce the number of strains being tested.
One way to do this is to use the information hidden in the genetic code of a strain to discover potential probiotic functionalities. Whole genome sequencing has become a cost effective and readily available technique in recent years. You can combine the genome information of bacteria with bioinformatics and computer modelling to very quickly screen large numbers of strains for the absence or presence of particular genes. And you can do this both for genes associated with a benefit, such as producing a specific vitamin, or for negative traits, such as transferable antibiotic resistance.
Taking that earlier example of potentially probiotic LAB strains, you look for genetic insight that they could potentially offer a similar health benefit as LGG or other known probiotic strains. You start with a pool of, say, 100 strains. You sequence their genomes, run them through computational predictive screening to evaluate the absence/presence patterns, and identify 10 or so that are most promising, based on the gene content. You can then design and carry out specific in vitro tests on 10 strains for the specific health benefit topic of interest, instead of 100 – which is already a big advantage in terms of cost and time. In addition, if they do show a probiotic effect in in vitro or even in vivo assays, the effect is substantiated with knowledge on the underlying mechanism, which could also support a potential patenting process.
RE: There are a number of standard bioinformatics tools that can be used to predict microbial functionality in silico. BLAST (basic local alignment search tool), for instance, compares the gene sequence of your potential probiotic to an existing database for a specific function or benefit, such as the production of short chain fatty acids. If your probiotic matches the references in the database, it may offer that functionality. This technique works well when you are comparing closely related species, but is not as good for distantly related species.
Another option that can be more widely applied and offer more reliable predictions is profile Hidden Markov Models (pHMMs). These are mathematical patterns that can be used to predict specific functionalities of unknown bacteria, even in distantly related species. Because they can also identify “patterns” or “motifs” in genes, they can highlight potential candidates that would be completely missed with traditional database comparison approaches. With pHMMs, you can go beyond the existing gene sequence databases and create customised profiles that target sets of specific functionalities and benefits.
RE: At NIZO, for example, we start with the information already available and published on the genes to identify those linked to the health benefit we are looking for: sleep quality, for instance. Often there will be a lot of different functionalities associated with a health benefit; in that case, we will make multiple pHMMs for a single health benefit, combined in a computational module. We then run the genome data for each candidate strain through the pHMMs, interpret the results, and classify each strain and its properties in its own “passport”.
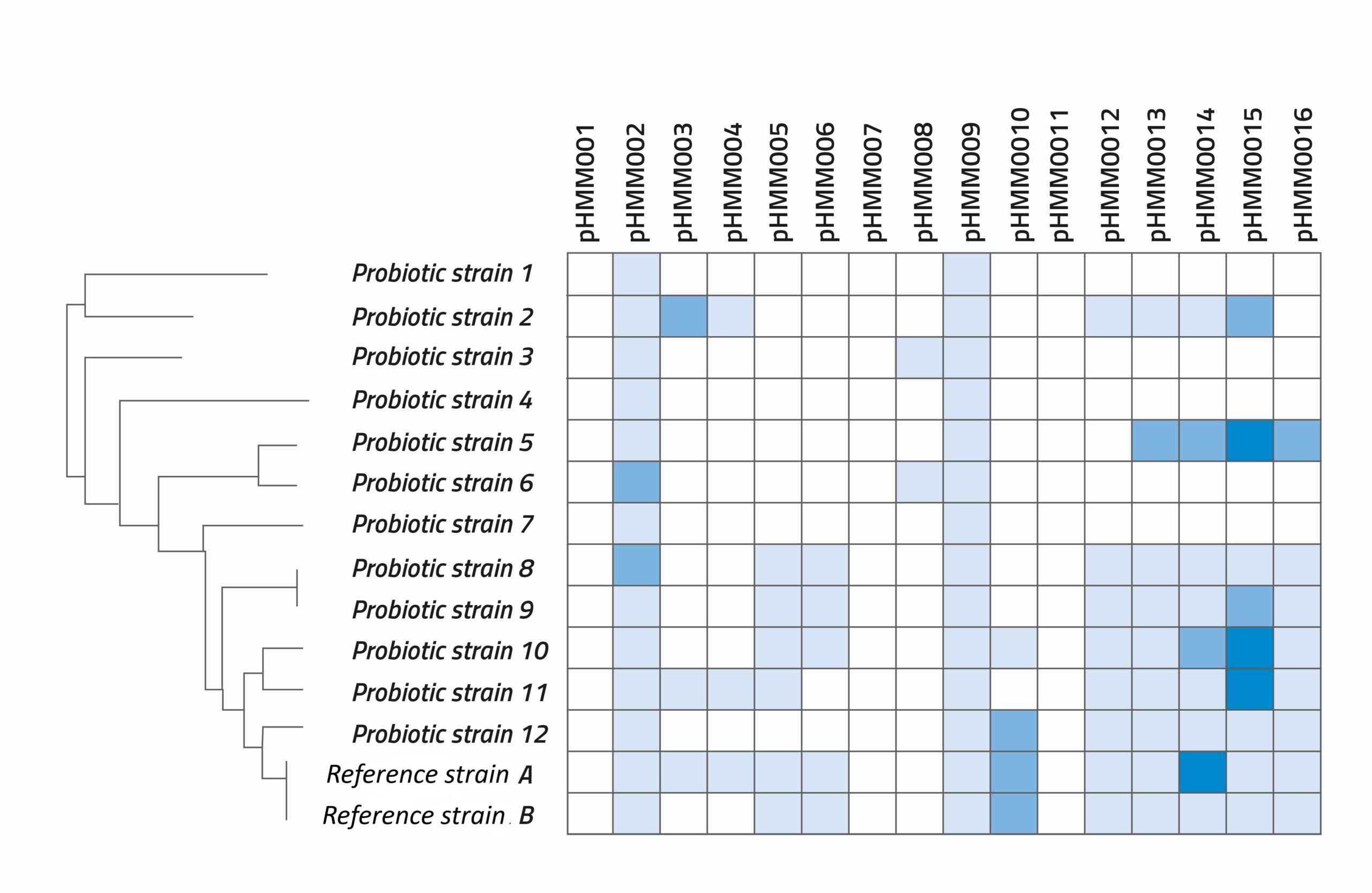
A key strength of this approach is that we can run multiple modules for different or related health benefits, and then create a matrix that includes the positive hit scores from them all. This overview allows bacterial strains to be compared in a more customised and complex way, to see which bacteria score high on some functionalities and low on others.
We have carried out projects looking into specific functionalities such as adhesion, production of short chain fatty acids, bile acid metabolism, etc. But we have also carried out more complex studies: on sleep quality, anxiety and stress, and skin health, for example. These projects have demonstrated that pHMMs are very suitable for investigating which types of probiotic strains may be beneficial for such health benefits.
RE: it is important to keep in mind that these are only predictions, and that they must be verified in vitro and in vivo (i.e., clinical trials) before a specific probiotic can be commercialised. There are so many interactions and other influences that will impact the actual behaviour of the probiotic, and the health impact it has.